AutoKinship is a Tier 1 tool at GEDmatch. Unlike Autotree (1), it does not require GEDcoms to be useful. AutoKinship creates possible family trees based on the amounts of DNA that the matches share with each other, which means that it doesn’t require any of those matches to have trees associated with their accounts.
Like most tools, all you have to do is input the kit number you’re using and adjust the settings.
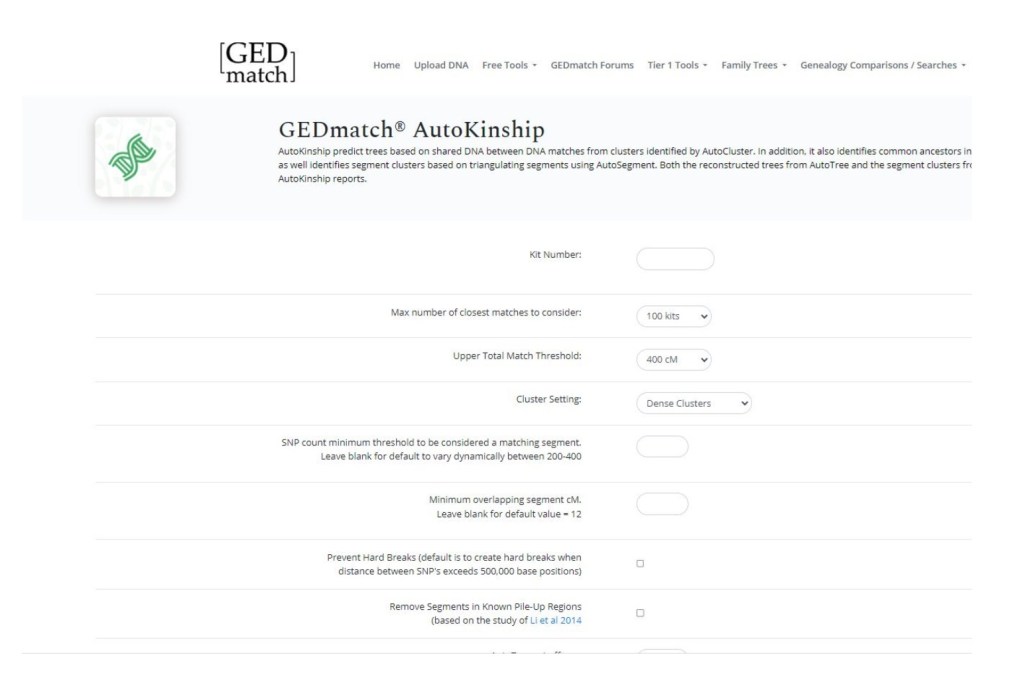
Once you submit you’ll be able to download the file.

Just like the AutoSegment tool, you’ll want to extract the zip file entirely out of the downloads folder. Once you open the folder, you’ll see the AutoKinship html file, which is what you want to open.

The AutoKinship file starts off with the now familiar clusters, followed by table of clusters with links for each cluster for AutoTree, ancestor names, and locations (if available), AutoKinship, AutoSegment (5), and surnames.
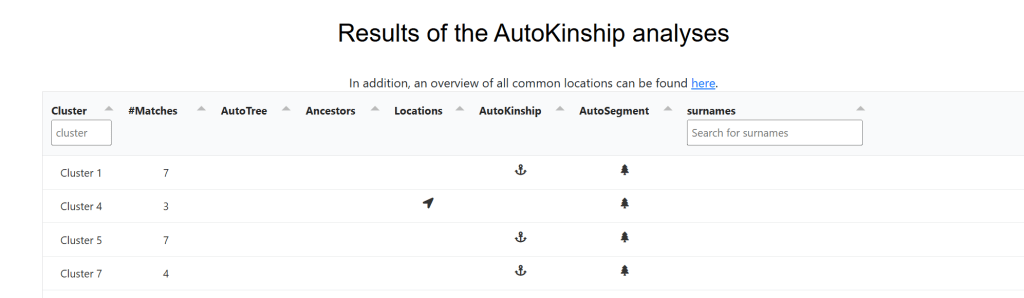
The AutoKinship analysis is followed by the a table with all the information on each match in each cluster, so you can see the names of the matches and how much DNA they share, and if they have a tree. If a particular cluster looks like it might be worth exploring, the AutoTree results may be helpful to see if you can find the common ancestor between yourself and your matches, or between your matches. However, since AutoTree requires your matches to have GEDcoms, this is not always an option. Enter AutoKinship.
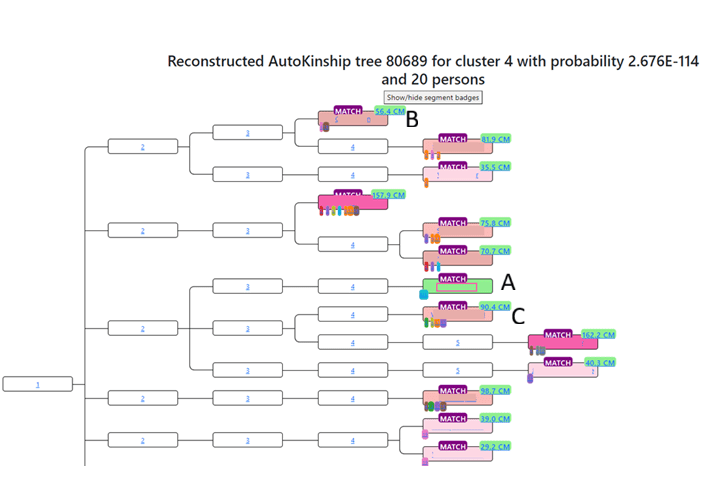
When you click on the AutoKinship link, it opens up a family tree, except the only names are the names of the matches. Each generation is labeled with their distance from the MRCA. The MRCA is 1, their children 2, their grandchildren 3, etc. shows how all the matches might be connected to the common ancestor. Each match has a “segment badge” that you can click on that will give you the names of the other matches who share that segment, and on which chromosome the segment they share is.
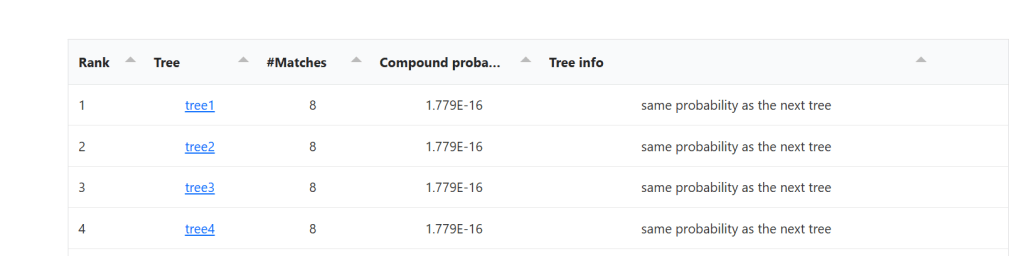
Underneath the tree is a ranked list of trees, and the tree info compares the probability of that tree being the correct tree as compared to the tree below it. The first tree is the same as the tree at the top of the page, but if you click on it, it will open in a new tab.
Underneath the ranked list, you will see a matrix which shows the amount of DNA each match shares with every other match. In addition, you can download a relationship matrix. Another way to see the relationship matrix is to open the tree in a new tab from the ranked list.
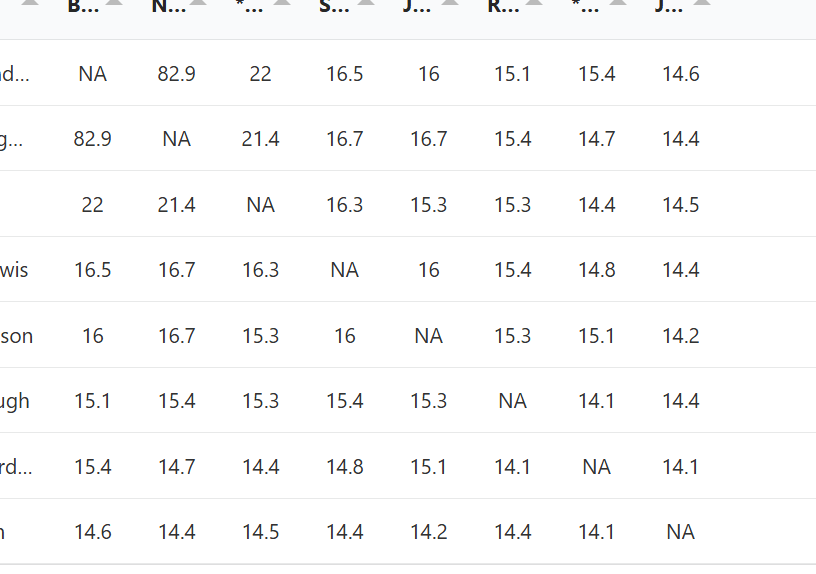
Instead of showing the amount of DNA between matches, the relationship matrix shows probability of the relationships between the matches as they are placed in the tree.
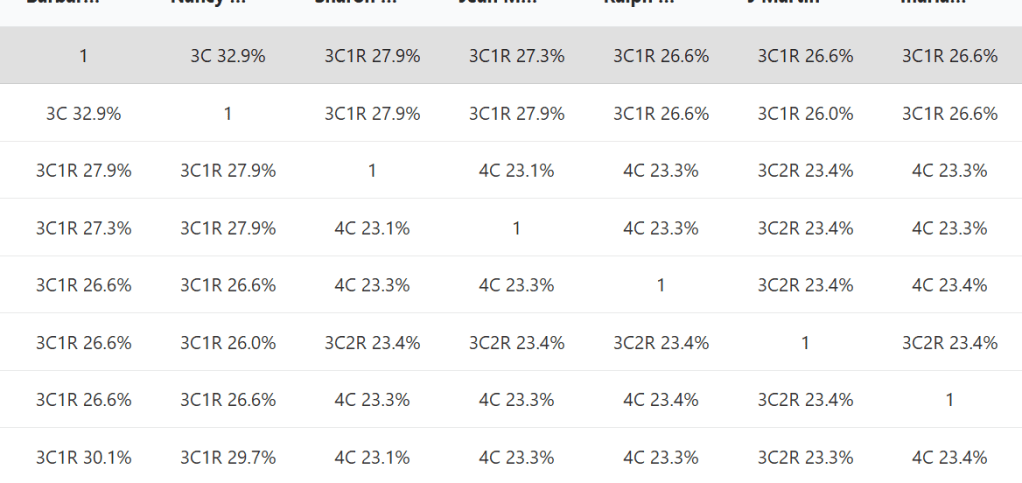
Clicking on the other trees from the ranked list will open up a similar tab, just with a slightly different tree and matrix which has relationships based on that tree.
Underneath the DNA matrix is the segment information of all the matches in that cluster, à la AutoSegment.
Let’s compare the above tree that AutoKinship created with the actual tree, since I know the relationship between some of the matches in this tree.
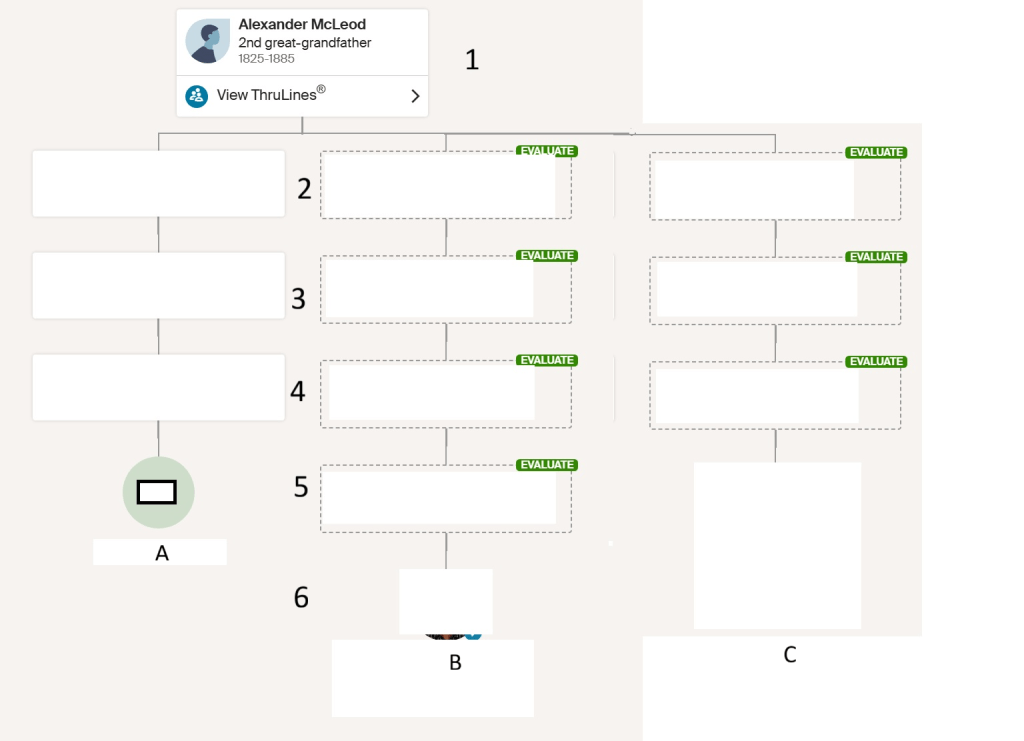
In the AutoKinship Tree, Match A and C are in generation 5, and match B is in generation 4. Match A and B were given the relationship of second cousin once removed (2C1R). Match A and C were given the relationship of second cousin (2C). Match B and C were given the relationship of second cousin once removed (2C1R).
In reality, match A and C are in generation 5, and match B is in generation 6. Match A and B have the relationship of third cousin once removed (3C1R). Match A and C have the relationship of third cousin (3C). Match B and C have the relationship of third cousin once removed (3C1R).
In another AutoKinship tree, these two matches are placed together as if they were second cousins (2C). Given the amount of DNA they share, this makes sense. They are placed in the same generation for every single AutoKinship tree created for this kit.
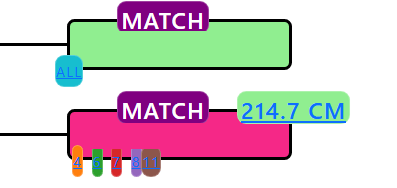
The reality, however, is that they are first cousins two times removed (1C2R). This means that none of the trees that AutoKinship proposed will be correct.
AutoKinship does provide a disclaimer that “there is no guarantee that all possible common ancestors are correctly identified,” and that it is possible to do a manual AutoKinship analysis in which you can define the generational information. But that’s a blog post for another day.
I can see why people enjoy the AutoKinship tree visualizations. They offer a quick way to imagine how a group of matches might fit together, and sometimes that spark of possibility is helpful. At the same time, because the trees represent hypothetical arrangements, I’m not sure the AutoKinship trees should be a part of the FIGG or Search Angel workflow. In those settings, spending time evaluating trees that might not reflect real relationships can pull focus away from the steps that actually move a case forward, like identifying genetic networks and documenting lines back to an MRCA, which you will still need to do even if the AutoKinship tree is correct. For me, the real value of the AutoKinship tool is everything bundled with it: AutoTree, AutoSegment, clustering, and the different matrices. And it’s reassuring that assembling the results of these tools into a complete picture still requires human reasoning, which automation can’t yet replicate.
- Jennifer Wiebe, “AutoTree at GEDmatch,” Jennealogie, 27 Nov 2025 (https://jennealogie.com/2025/11/27/autotree-at-gedmatch/).
- Jennifer Wiebe, AutoKinship settings, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Download results, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Contents of Autokinship folder, 2 Dec 2025, author’s files.
- Jennifer Wiebe, “AutoSegment at GEDmatch,” Jennealogie, 10 Nov 2025 (https://jennealogie.com/2025/11/10/autosegment-at-gedmatch/).
- Jennifer Wiebe, AutoKinship analysis, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Sample AutoKinship Tree, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Ranked tree list, 2 Dec 2025, author’s files.
- Jennifer Wiebe, DNA Matrix, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Relationship matrix, 2 Dec 2025, author’s files.
- Jennifer Wiebe, AutoKinship comparison tree, 2 Dec 2025, author’s files.
- Jennifer Wiebe, Sample AutoKinship Tree 2, 2 Dec 2025, author’s files.

Leave a reply to geneticaffairs Cancel reply