Now that I’ve explained the clustering (1)and triangulation (2)tools at GEDmatch, the scene is set to introduce a tool that I think of as the baby of these two tools.
AutoSegment is a Tier 1 tool which you can find in the sidebar or dropdown menu. You enter a kit number and the number of matches you want to use, and much like the clustering tool you can specify an upper and lower threshold. In addition, you can set a minimum segment threshold. The default is at 10, but if you only wanted to deal with bigger segments you can raise it. The threshold can’t be below 7, for good reason.(3) You can also choose whether you want to look at all chromosomes or just one in particular.
Roberta Estes, (4) who is far more knowledgeable than me, suggests using a minimum of 500 SNPs to increase the likelihood of Identical by Descent segments (I wrote about what IBD means here (5)).
In terms of “prevent hard breaks,” what is important to know is that the companies are not using all the same SNPs for their tests. While there is a lot of overlap, some test SNPs that others don’t, and vice versa. That means when they all come to GEDmatch to be compared, there’s going to be some gaps. Since we calculate segments by a string of matching SNPs, if one SNP is missing (because one company tested it and the other didn’t), a bigger segment is going to be broken down into two segments, and if both of them are now too small to be included, that match will be excluded. By preventing hard breaks, as long as there is a matching SNP on either side of the missing SNP, this won’t happen.
If you want to avoid known pile-up regions,(6) you can select that box. Run the tool twice to see if you notice a difference if it’s selected!

Once you hit submit, you will be taken to a page where it will think for a little bit before you will be able to download your results. The results come in a zip file so if your computer doesn’t unzip it automatically, you will have to unzip it first. Save the file somewhere to your computer. If you try to run it from the downloads folder, you’ll get errors when you click on links in the HTML file. There are two AutoSegment files, one bigger (with the chart) and one smaller (with no chart).

Upon opening the bigger HTML file, you will be taken to a page that looks very familiar. Just like in the clustering tool, the small colourful boxes will slide around until you have many different coloured clusters.
If you scroll past that box, you’ll first see the “Settings used for this AutoSegment analysis” which explains to you how many segment clusters, matches, and segments were found.

Don’t confuse Segment Cluster with Cluster. The DNA matches were sorted into 48 multi-coloured clusters, but within those groups, you might have more than one group of segments each cluster shares. This says that there are 69 segment clusters.
There’s also a small section about chromosome browsers, (10) (and how to import this information to DNA Painter), followed by the segment statistics per cluster. Scroll down further to get to the AutoSegment Cluster Information, followed by Individual segment Cluster Information, which I will talk about in more detail below.
Before we get into these sections, let’s talk about how the AutoSegment clustering is different than the regular clustering. Regular clustering is creating genetic networks by shared matching. AutoSegment is creating genetic networks using triangulated segments (overlapping segments on the same maternal or paternal chromosome). This is why I think of this tool as the baby of the clustering tool and the triangulation tool.
Here is the difference between the clustering tool and the AutoSegment tool with similar settings.
In the clustering tool, I have a match named A, who appears in this green cluster:
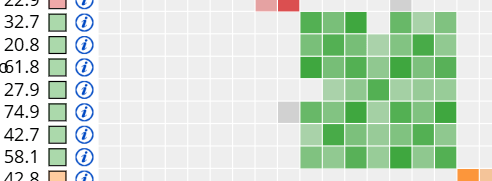
Here is that same match in the red AutoSegment cluster:

All matches in a genetic network will not triangulate, so when you’re using triangulation to create your genetic networks, you are creating a subset of the members of the original genetic network. We like triangulated matches because we know they share a common ancestor that they all inherited that shared segment from, whereas non-triangulated matches in a genetic network only hypothetically share a common ancestor. However, it’s always best to work with as many matches as possible, so while the triangulated matches give weight to the genetic network, we don’t want to discard other non-triangulating matches. Triangulate, but don’t eliminate.
AutoSegment Cluster Information

This table, grouped by cluster, is going to tell you who is in each of your clusters, their email, the cluster number, how much DNA they share, and the number of shared matches.
If you click on any person, you’ll get a report. This is a different way of seeing the information, because sometimes matches share more than one triangulated segment.

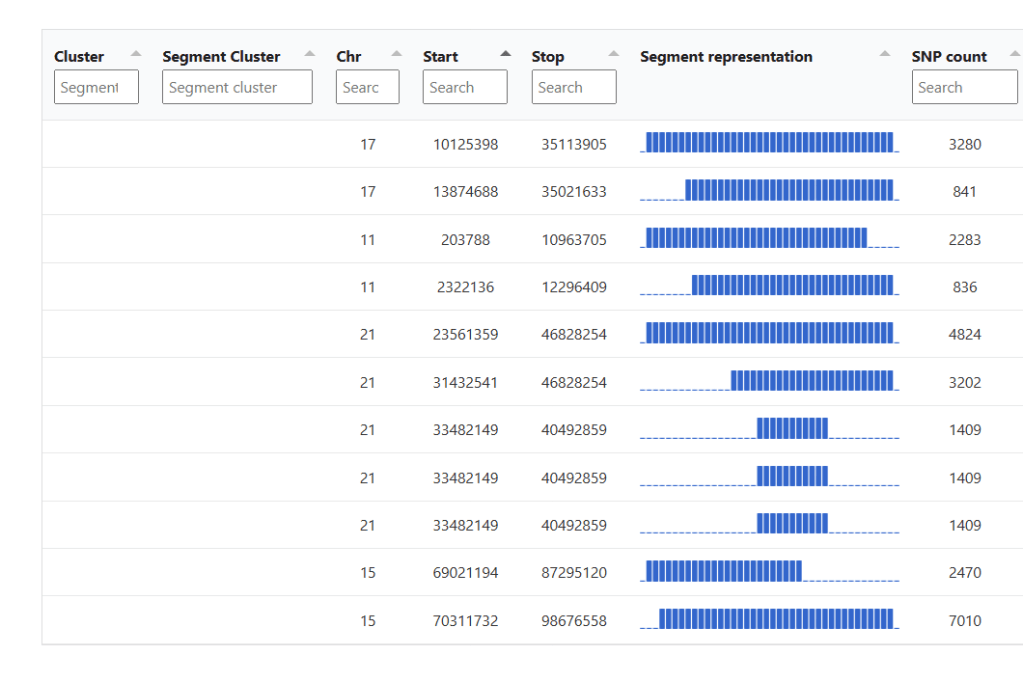
You’ll also get to see those segments in a chromosome browser:

Since this match shares segments with other matches in more than one place, the DNA match report reclusters this smaller group of matches, and also provides an autocluster cluster information to explain the clusters. That chart will include whether or not any of the matches have trees.

This is followed by the segment clusters for that match, which visualizes the shared segments. Note that four different chromosomes are represented here.
Individual segment Cluster Information
This is the similar as the above image, but will all the segment clusters instead of just one DNA match’s segment clusters. It is grouped by segment cluster instead of by cluster.
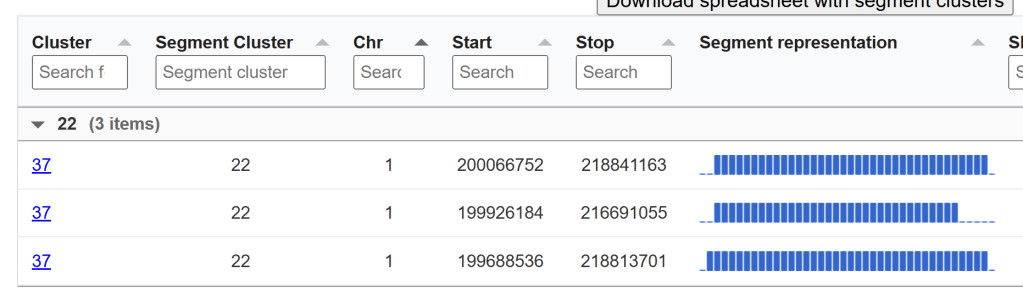
In this next image, you can see that both of these groups are part of cluster 7, but their segment cluster group number is different. One is segment cluster group 6, and one is segment cluster group 32. It’s entirely possible that they should all be part of the same segment group, but because of the settings (perhaps one person shared less than 10cM overlap with another person), they got put in separate segment groups. They do all cluster together, however.

I hope it’s obvious why I think of this tool as the baby of autocluster and triangulation. We have the organization of clusters (which the triangulation tool lacks) along with the certainty of the confirmed triangulated segments (which the clustering tool lacks). So how can this help us with our research? We know that the triangulated segments guarantee that everyone in the cluster shares a common ancestor, which means our next focus is to try to find out who that common ancestor is.
If you’re a genealogist researching a kit with known DNA matches, the DNA matches that triangulate with those known matches are going to share a common ancestor somewhere on line that you share with the known DNA matches. For example, if my second cousin Andrew, with whom I share great-grandparents Belinda and Carl, triangulates with matches Daniel and Evan, I know that I’m going to find the common ancestor all of us share on Belinda or Carl’s line. Having as many people from all different branches of your family tested will allow you to put all the triangulation groups into different buckets representing different branches of your family tree, making it easier to figure out who that common ancestor is.
If we don’t have any known DNA matches (which is always the case if we are working on the kit of someone who is adopted), we can still mine the clusters for clues. DNA matches who have trees will be really helpful, but even without trees we can try to find out as much information as we can about these matches, such as such as shared ethnicity, locations, or surnames.
At first glance, the AutoSegment tool is overwhelming. Despite the amount of information presented, the big picture should not be forgotten. The triangulated clusters are the best version of a genetic network, which means that instead of exploring your match list one by one, these super genetic networks let you harness the power of multiple matches all at once.
- Jennifer Wiebe, “Clustering at GEDmatch,” Jennealogie, 8 Nov 2025 (https://jennealogie.com/2025/11/08/clustering-at-gedmatch/).
- Jennifer Wiebe, “Triangulation, Part 5,” Jennealogie, March 9, 2024 (https://jennealogie.com/2024/03/09/triangulation-part-5/).
- Jennifer Wiebe, “Don’t touch the settings!,” Jennealogie, April 20, 2018 (https://jennealogie.com/2018/04/20/dont-touch-the-settings/).
- Roberta Estes, “AutoSegment Triangulation Cluster Tool at GEDmatch,” DNAeXplained, 18 Oct 2021, (https://dna-explained.com/2021/10/18/autosegment-triangulation-cluster-tool-at-gedmatch/).
- Jennifer Wiebe, “DNA in a Nutshell,” Jennealogie, February 13, 2018 (https://jennealogie.com/2018/02/13/dna-in-a-nutshell/).
- Jennifer Wiebe, “Triangulation, Part 4: Pileups,” Jennealogie, March 19, 2018 (https://jennealogie.com/2018/03/19/triangulation-part-4-pileups/).
- Jennifer Wiebe, AutoSegment settings, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Download AutoSegment results, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Settings for AutoSegment analysis, 10 Nov 2025, author’s files.
- Jennifer Wiebe, “What’s the use of a chromosome browser?,” Jennealogie, May 4, 2018 (https://jennealogie.com/2018/05/04/whats-the-use-of-a-chromosome-browser/).
- Jennifer Wiebe, AutoClustering close up, 10 Nov 2025, author’s files.
- Jennifer Wiebe, AutoSegment Cluster close up, 10 Nov 2025, author’s files.
- Jennifer Wiebe, AutoSegment Cluster Information, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Information about DNA match, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Chromosome Segments, 10 Nov 2025, author’s files.
- Jennifer Wiebe, DNA match cluster, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Segment Clusters for DNA match, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Individual Segment Cluster Information, 10 Nov 2025, author’s files.
- Jennifer Wiebe, Individual Segment Cluster Information 2, 10 Nov 2025, author’s files.

Leave a reply to maltsoda Cancel reply