There’s nothing that makes you feel older than when you start talking about how you used to do things “back in the day.” The problem with genetic genealogy is that back in the day isn’t really that far back. Since the advent of consumer DNA testing 25 years ago, great strides have been made to help us understand and make use of our DNA. Since GEDmatch’s founding in 2010, they have added a significant number of tools for that purpose. In 2017 Jonny Perl offered us DNA Painter chromosome mapping (1), the first of many tools in that suite. In 2018, Dana Leeds published the Leeds method (2) to manually cluster your matches, followed by EJ Blom’s Genetic Affairs tool (3), which automated it. That same year, the field entered a new era with the arrest of the Golden State Killer (4). All these things solidified the need for a genetic genealogy workflow.
As the range of available tools has grown, some of the methods we used to use have changed. I’ve already written about how we used to do (5) triangulation on GEDmatch and how the advent of the Triangulation tool (6) now simplifies that process. The AutoSegment Split tool similarly streamlines things.
Before we dive into how it simplifies our workflow, let’s take a look at what the tool actually does. The settings for this tool are similar to the settings for many others, except that it adds the selection of an Admixture project (7). More on that later.

Once the tool runs, you will be able to download the files. Don’t forget to extract the zip file to another folder!

The AutoSegmentOppositeSides html file is the one you’re looking to open.

AutoSegment Split starts off just like the AutoSegment tool (11). You have matches grouped into clusters, and their segment detail.
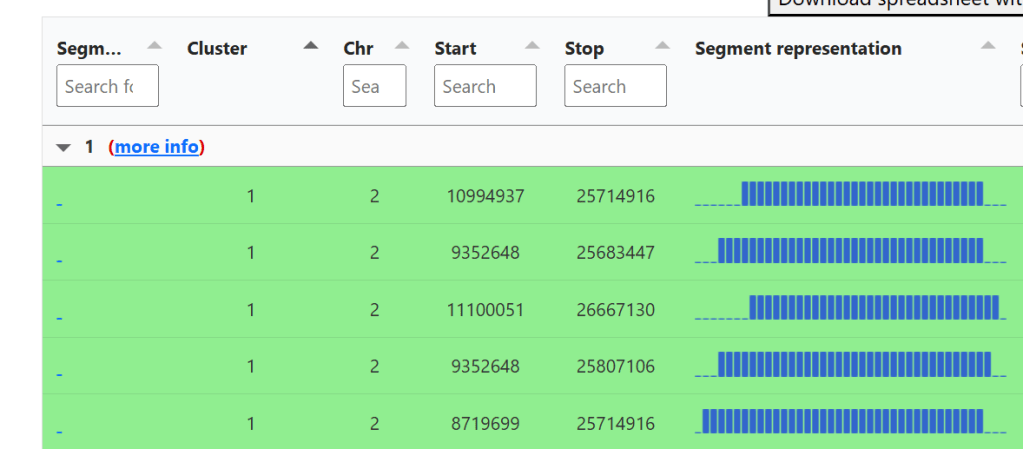
Clicking “More info” opens a page with a matrix that lists every match in the cluster and shows the size of the shared segments between each pair.

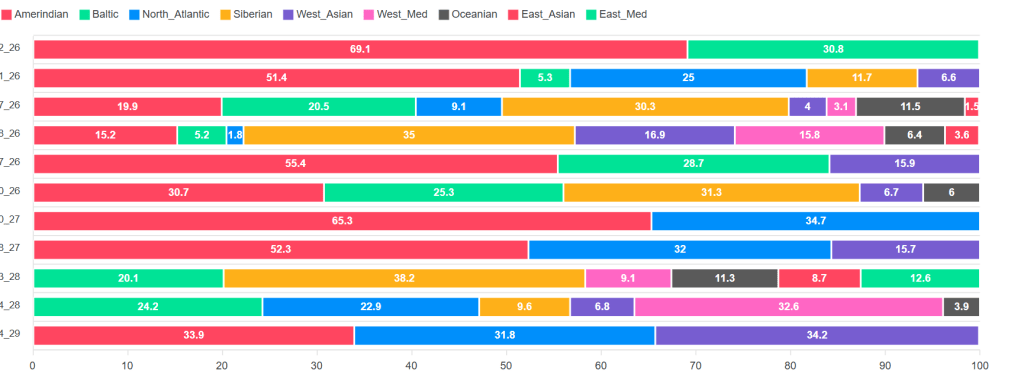
After the matrix we can see the Admixture results for that segment. Remember how we had to input an Admixture project? That’s for this part of the tool. The original Admixture tool offers a broad overview of a kit’s ethnicity estimate. Some companies break that down, chromosome by chromosome. AutoSegment Split offers a segment by segment breakdown, which might help you pinpoint which branch of the family tree you’re working with, if the ethnicity of that branch happens to be distinct from the other branches. In the following example, I can see that the shared segment likely came from one of my Indigenous ancestors.
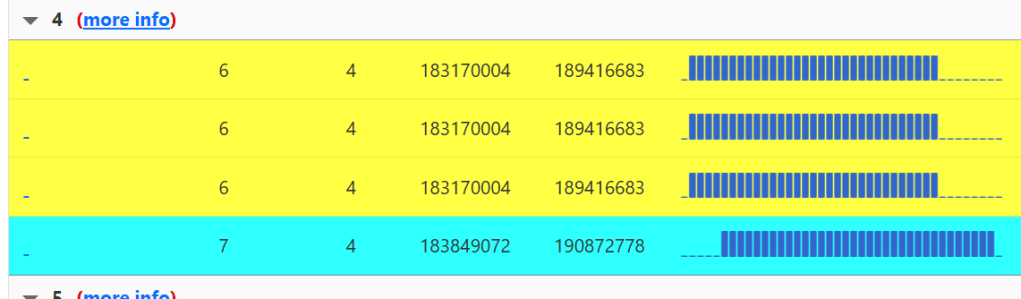
Besides segment admixture, another advantage that this tool offers over the regular AutoSegment is that in some clusters, you’ll have more than one colour.
Here’s where I have to start with “back in my day” so you can appreciate how this simplifies the workflow. Triangulation was as useful to us then as it is now, because we know for sure that triangulated matches share a common ancestor. A triangulation exists when matches share an overlapping segment on the same chromosome. We do have to be careful here because we’re not just talking about the chromosome number being the same. In addition to the chromosome number being the same, since each chromosome number represents a pair of chromosomes, the segments have to overlap on the same maternal or paternal chromosome to triangulate. This is easy to visualize using the DNA Painter chromosome mapping tool.
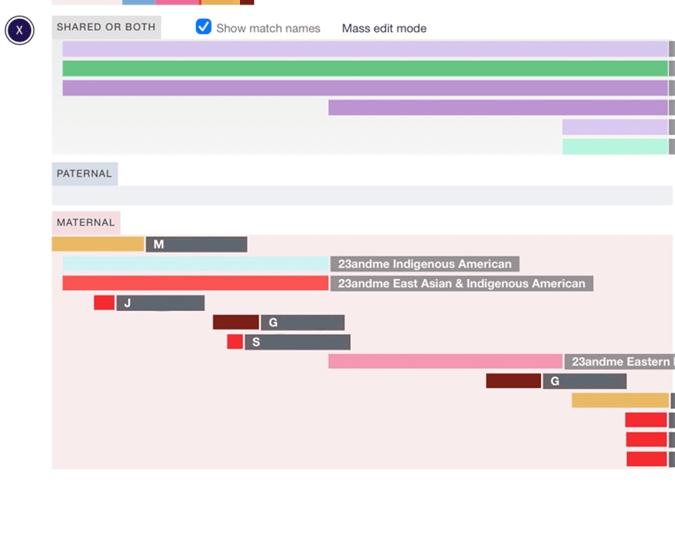
In this map, the purple and greenish segments represented in the “shared or both” section on the X chromosome overlap, which means they may triangulate. We can’t tell for sure because they haven’t been assigned to the maternal or paternal X chromosome. The segments that overlap on the maternal section of the X chromosome on this map definitely triangulate.
What if we ran the segments that we’re not sure of through the triangulation tool? Imagine that we did that, and the purple segments triangulated with each other, but not with the greenish ones, and the greenish ones triangulated with each other, but not with the purple ones. This tells us something! If we have two different triangulation groups in the same spot, they each represent a different chromosome, maternal or paternal. If we can’t identify which is maternal or which is paternal, we can just refer to them as Parent A and Parent B. Knowing that these two groups are on different sides of the family tree can be helpful in our research.
Instead of having to do all that work to assign parental sides, AutoSegment Split does the work for us. In the yellow and blue cluster above, the segments all overlap on chromosome 4. Two different colours are used because the blue segment doesn’t triangulate with the yellow ones. That means that blue is on one side of the family tree, and yellow is on the other. We can see it very clearly when we look at the segment matrix for this cluster. Blue doesn’t share any DNA with yellow.
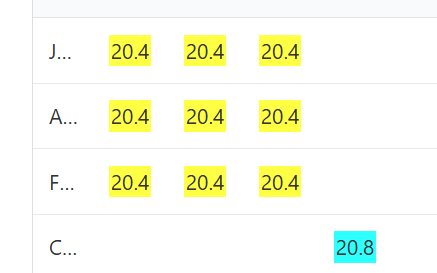
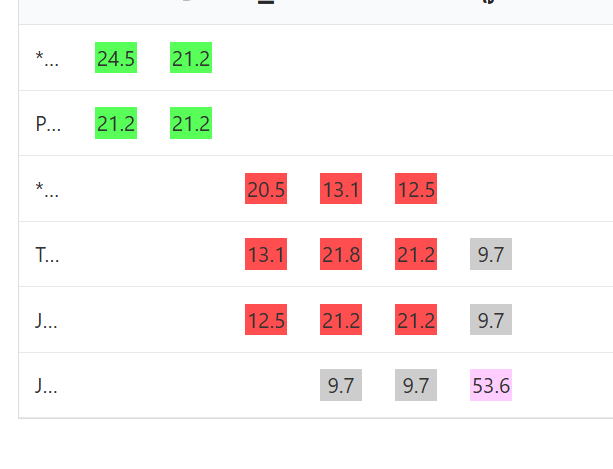
It can happen that sometimes more than two colours will be assigned to a segment cluster.

Red does not triangulate with green, but there’s enough overlap between the two that we can say these two colours are on opposite sides of the family tree. Pink does not overlap at all with green, so there can’t be a triangulation there, but it also doesn’t overlap enough with red, so we can’t really place pink on the same side as red or green. If we look at the matrix, we can see that pink probably does belong with red, it’s just not sharing large enough segments with the people in the red group, including one match that it doesn’t share a segment with at all. I suspect that the segment they share is so small that the tool is ignoring it altogether.
Now that we’ve seen how the tool works, the next question becomes clear: how do we integrate it into an overall workflow? As I mentioned in my post on the AutoKinship tool (20), my work is in the framework of Search Angel or FIGG casework, taking a GEDmatch kit from unknown to identified. A big part of that workflow is finding genetic networks. (21) Once we do that, we build back the trees of our network, find the MRCA between the matches, and then build down to identify the DNA.
With that workflow in mind, deciding whether or not a tool is worth adding in, I first consider whether it’s going to cut down the time it takes to currently do what the tool does. AutoSegment Split significantly cuts down the time it takes to determine whether overlapping segments are on opposite sides.
My next consideration is whether the tool provides probative information. Not every genetic network is going to be helpful for identifying unknown DNA. This process works best with matches that are fairly close. First, we want to avoid having to build our trees really far back to identify an MRCA, since many trees don’t go very far back. Second, even if we could identify that distant MRCA, building down from that point can involve navigating thousands of possible descendants. According to the ISOGG wiki, (22) a person may have as many as 17,300 fifth cousins.
For that reason, our focus is usually on matches at the second cousin level, but third or even fourth cousins can sometimes be helpful as well. Given that every genetic network is different, it’s impossible to come up with hard and fast rules to evaluate which genetic networks will be useful. No sooner than you hit enter on a comment stating that 20cM matches aren’t very probative, someone will reply to you that these matches shouldn’t be discounted because that’s how much DNA they share with a third cousin.
While I still need to refine my thinking about what constitutes a probative genetic network, I approach this question by considering two factors: the amount of DNA shared with the original kit, and the amount of DNA shared between the matches in the network. Since the AutoSegment Split tool only gives me the size of the segments that these matches share, I can’t easily evaluate how probative a particular cluster might be. I would have to take all these matches and manually enter them into the Multi-Kit analysis tool to create a matrix to see how much total DNA they share with each other. Doing so for every cluster to find one that is probative would be a lot of work.
To see how this would work in practice, I did this for the green-red-pink cluster. You saw that one of the matches shares a segment of 50cM, which is pretty significant! That match also shares a total of 477cM of DNA with the kit, but the other matches aren’t very significant to the kit, nor to each other.

Moving past the example of this cluster, it’s clear that the tool has practical uses in other areas of research. For example, when I already know the identity of the person behind the original kit and am trying to extend their ancestry further back, it can be very helpful. I can see that some of the matches have trees, and in one of the trees I can see someone has ancestors going back to a known ancestral area of this kit on the paternal side. This person is part of the red group, which means it’s likely that red is paternal, and green is maternal. If I was interested in pursuing a particular branch on either side of my tree, I would be looking into those matches, even if they are very distant. And as I mentioned above, the Admixture breakdown for each segment can also provide clues.
The Admixture breakdown for each segment can also be useful when working with unknown DNA. If many of your matches come from one branch of the family tree with a particular admixture, it can be difficult to identify a different branch to investigate. By using the Admixture tool to spot segments with a different admixture, you can prioritize clusters that might lead to new lines of inquiry.
Even so, as with most tools in genetic genealogy, some matches can appear on both sides of the family tree, and it’s important to be aware of that. For example, in my red-green-pink cluster, the original kit is my great-aunt, and the significant match is me. It can be challenging to determine sides since I appear in clusters on both her maternal and her paternal sides. This doesn’t even take into account endogamy, where matches are more likely to link to multiple branches of the family tree. For instance, my father has a fourth- and fifth-cousin (the same person is related in three different ways) who shares common ancestors on both his maternal and paternal sides.
Challenges like that highlight that no tool, by itself, can provide all the answers. While the AutoSegment Split tool simplifies much of the workflow, understanding how a tool works and how to interpret its output is only part of the picture. The real key is understanding what your research goal is and how the results from any tool will help you move toward it. When the goal and the tool align, even simple outputs can be powerful. When they don’t, even the most sophisticated automation won’t get you closer to an answer.
- DNA Painter (https://dnapainter.com/).
- “The Leeds Method,” Dana Leeds (https://www.danaleeds.com/the-leeds-method/).
- Genetic Affairs (https://geneticaffairs.com/)
- Jennifer Wiebe, “GSK, 5 years later: Still building the plane,” Jennealogie, April 20, 2023 (https://jennealogie.com/2023/04/20/gsk-5-years-later-still-building-the-plane/).
- Jennifer Wiebe, “Triangulation,” Jennealogie, February 24, 2018 (https://jennealogie.com/2018/02/24/triangulation/).
- Jennifer Wiebe, “Triangulation, Part 5” Jennealogie, March 9, 2024 (https://jennealogie.com/2024/03/09/triangulation-part-5/).
- Jennifer Wiebe, “Gedmatch Basics, Part 1,” Jennealogie,
March 1, 2018 (https://jennealogie.com/2018/03/01/gedmatch-basics-part-1-2/). - Jennifer Wiebe, AutoSegment Split Settings, 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Split download , 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Split folder contents, 9 Dec 2025, author’s files.
- Jennifer Wiebe, “AutoSegment at GEDmatch,” Jennealogie, February 24, 2018 (https://jennealogie.com/2018/02/24/triangulation/).AutoSegment blog post
- Jennifer Wiebe, AutoSegment Split cluster, 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Split segment matrix, 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Segment Ethnicity, 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Cluster Colours, 9 Dec 2025, author’s files.
- Jennifer Wiebe, DNA Painter chromosome map, 9 Dec 2025, author’s files.
- Jennifer Wiebe, Segment Matrix, 9 Dec 2025, author’s files.
- Jennifer Wiebe, AutoSegment Cluster with more than 2 colours, 9 Dec 2025, author’s files.
- Jennifer Wiebe, Segment Matrix 2, 9 Dec 2025, author’s files.
- Jennifer Wiebe, “AutoKinship,” Jennealogie, December 2, 2025
(https://jennealogie.com/2025/12/02/autokinship/). - Jennifer Wiebe, “What is a genetic network?,” Jennealogie, July 29, 2024,(https://jennealogie.com/2024/07/29/what-is-a-genetic-network/).
- “Cousin statistics,” ISOGG Wiki, last edited 27 February 2022 (https://isogg.org/wiki/Cousin_statistics).
- Jennifer Wiebe, Matrix of matches, 9 Dec 2025, author’s files.

Leave a comment