A new tool recently came out at GEDmatch, and I wanted write a blog post about it so I could understand it better. I realized, however, that I had never written about GEDmatch’s clustering tool, which is useful for understanding how this new tool works.
I’ve talked about clustering (1) before, when it was a stand-alone tool that existed on its own webpage. Now, the same tool exists at GEDmatch, making it easier than ever to group your matches into genetic networks. (2)
Clustering is a Tier 1 tool at GEDmatch that can be accessed using the sidebar or top drop-down menu (anyone else always forget about the drop-down menu because you’ve been using GEDmatch since before it existed?). Note that in the drop-down menu it is called AutoTree Clustering, whereas in the side bar it is called Clusters with AutoTree, Closest to Single Kit version, but regardless of which one you click it will take you to the same tool: “Closest Matches AutoCluster – AutoTree – AutoPedigree.”

Now all you need to do is pop in your kit number and adjust the settings to your liking. I like to raise the number of kits, because that will likely lead to more and/or bigger clusters. In terms of the centiMorgans threshold, the lower threshold is the minimum amount of centiMorgans that a match must have to be considered. The upper threshold is the highest amount of centiMorgans a match can have. I don’t change the overlap, nor do I include segment detail, and I’ll briefly talk about Auto Tree later. The cut off year has to do with the Auto Tree feature.
Once we hit the cluster button, we wait until it’s done thinking and hit display clusters:

Then a big grey box with a bunch of colourful boxes will appear, followed by everyone’s favourite part, the coloured boxes sliding around until they are grouped with other boxes of the same colour. We end up with coloured boxes made up of smaller boxes. These are your clusters. Each of these clusters represents a genetic network, and hypothetically, everyone who is the same colour can be traced back to a common ancestor.

You’ll also notice some random smaller grey boxes. These boxes show overlap between two clusters: people in one cluster also share DNA with people in another cluster. Too much overlap means the clustering tool won’t be useful, because we want to have distinct genetic networks. Usually when too much overlap is an issue, you’ll end up with one giant cluster and a bunch of really small ones, with a ton of small grey boxes. Here the overlap is very mild.
You can see that the matches in each cluster share around 20cM with each other, and comparing the list of clustering matches to the one-to-many match list, I can see that some of this kit’s highest matches weren’t included.
Sometimes those matches don’t have any other matches who are part of their genetic network. Sometimes, this happens because of the way we have adjusted our settings, so try playing around with them and see what happens. Here I raised the lower threshold to 30 and I raised the upper threshold to 500. If you remember the Leeds Method (6), you want to include matches as high as 400cM, and 500 is the closest setting available to that. Now we have some pretty useful clusters. The closer our matches are, the easier they are to work with, so in terms of promising clusters, red and grey would be where I would want to start.
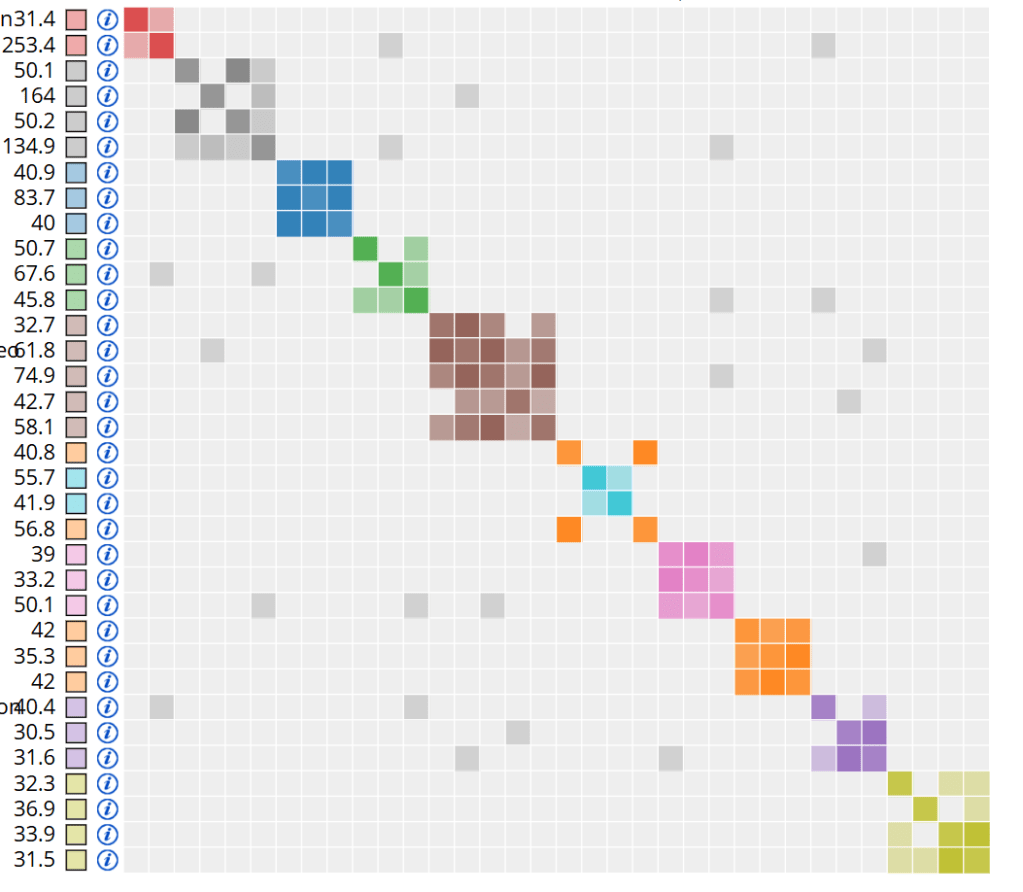
Each of the boxes next to the amount of centiMorgans can be checked off and then submit to Multi-Kit-Analysis. I like to run the matrix (8) tool for each of my clusters to see how the matches in that cluster match each other.

You can also do clustering from the one-to-many tool. From the one to many list, select the matches you want to include and click on visualization options:

The clustering tab is the one on the far right:
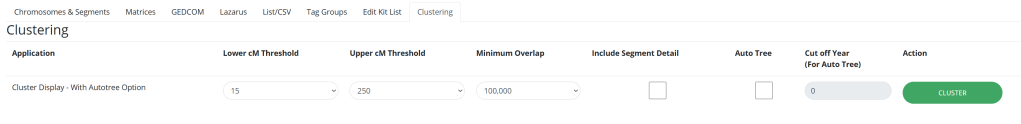
Again, we can adjust the settings to our liking before hitting the clustering button.
If you select Auto Tree, it will look at the GEDcoms (family tree files) of the matches in each cluster, and see if it can find any common ancestors between them. The year is how far back in the pedigree you want to go, and the default is 1700.

If Auto Tree is selected, there will be a few extra tabs to look at. The Auto Tree AutoCluster Analysis tab is where you’ll find out if there are common ancestors in any of the clusters. The symbols below will show up next to the cluster: the tree and arrow opens up the common ancestor and common location, whereas the paper will download the GEDcom file of those common ancestors.

If their common ancestor is not someone in your tree, this gives you a lead to follow up on. It’s possible that their common ancestor is also your common ancestor, or that the common ancestor between all of you is somewhere on that ancestor’s line. Understand that this is a hypothesis, so hold that information lightly. Also, while Auto Tree is a great tool that can cut down the time it takes to look through GEDcoms to find a common ancestor, the rest of the matches in the cluster that don’t have GEDcoms should be explored as well.
Seeing each genetic network grouped together in a cluster helps organize our research by allowing us to tackle smaller groups of matches rather than one long list of individual matches. Starting with the hypothesis that the members of a cluster all share a common ancestor (aided perhaps by Auto Tree), we can begin our research and follow those leads wherever they may take us.
- Jennifer Wiebe, “Clustering at Genetic Affairs,” Jennealogie, 24 January 2019 (https://jennealogie.com/2019/01/24/clustering-at-genetic-affairs/).
- Jennifer Wiebe, “What is a genetic network?,” Jennealogie, July 29, 2024,(https://jennealogie.com/2024/07/29/what-is-a-genetic-network/).
- Jennifer Wiebe, GEDmatch AutoCluster tool, 8 Nov 2025, author’s files.
- Jennifer Wiebe, GEDmatch Disply Clusters, 8 Nov 2025, author’s files.
- Jennifer Wiebe, GEDmatch Clusters 1, 8 Nov 2025, author’s files.
- Dana Leeds, “DNA Color Clustering: The Leeds Method for Easily Visualizing Matches,” Genealogy With Dana Leeds, 23 August 2018 (https://www.danaleeds.com/dna-color-clustering-the-leeds-method-for-easily-visualizing-matches/).
- Jennifer Wiebe, GEDmatch Clusters 2, 8 Nov 2025, author’s files.
- Jennifer Wiebe, “Welcome to the Matrix,” Jennealogie, 9 Dec 2023 (https://jennealogie.com/2023/12/09/welcome-to-the-matrix/).
- Jennifer Wiebe, Multi Kit Analysis, 8 Nov 2025, author’s files.
- Jennifer Wiebe, Visualization Options, 8 Nov 2025, author’s files.
- Jennifer Wiebe, Clustering Tab of Visualization options, 8 Nov 2025, author’s files.
- Jennifer Wiebe, AutoTree AutoCluster Analysis, 8 Nov 2025, author’s files.
- Jennifer Wiebe, AutoTree icons, 8 Nov 2025, author’s files.

Leave a comment