If you’re expecting to choose between a red and blue pill right now, you’re going to be sorely disappointed. But if you’re looking to understand how to read a matrix, you’re in the right place!
First, how do we create a matrix? This can be done easily in GEDmatch (1), regardless of whether or not you have tier 1 subscription.
First, go to “One-to-Many – Original Version”
You’ll be prompted to put in a kit number or select a kit from the pull down menu:
Once you have your results, select the kits you want to be included in your matrix (note that you can also do this in tier 1 one-to-many):
Once you’ve selected your kits, click “visualization options.” All the kits you’ve chosen are there, and you can manually add any other kits you want to include. Click “visualization options again” when you’re ready.
You’ll be taken to a page with lots of different options, but Matrices is the one we’re looking for:
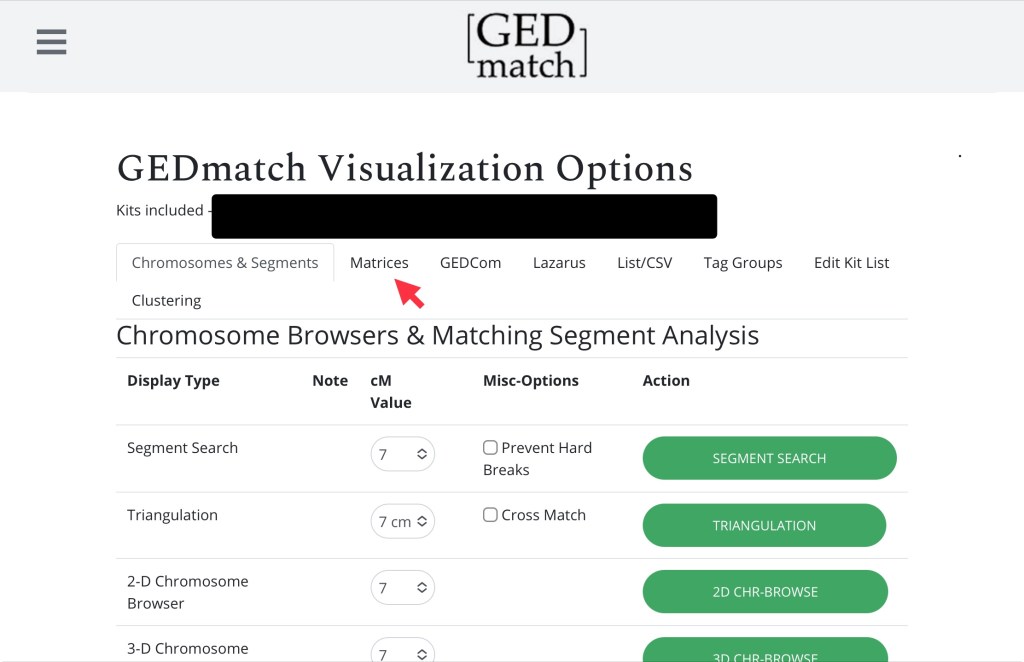
Here we’re just interested in an autosomal matrix. Do I need to remind you not to touch the settings (7)?
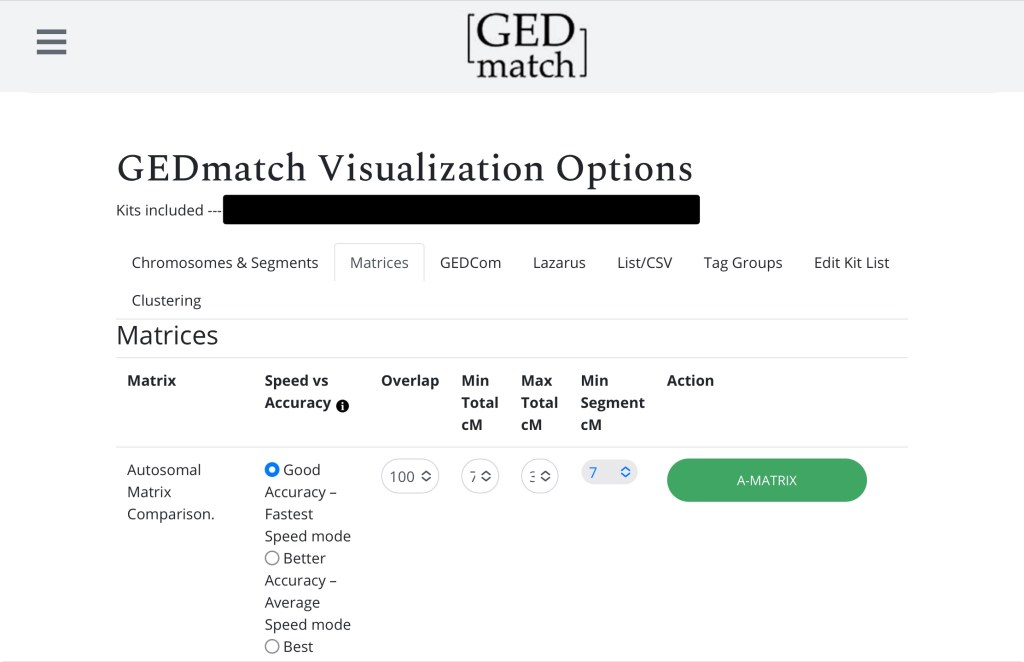
Click A-Matrix and voilà! You have a matrix that looks a little like this:
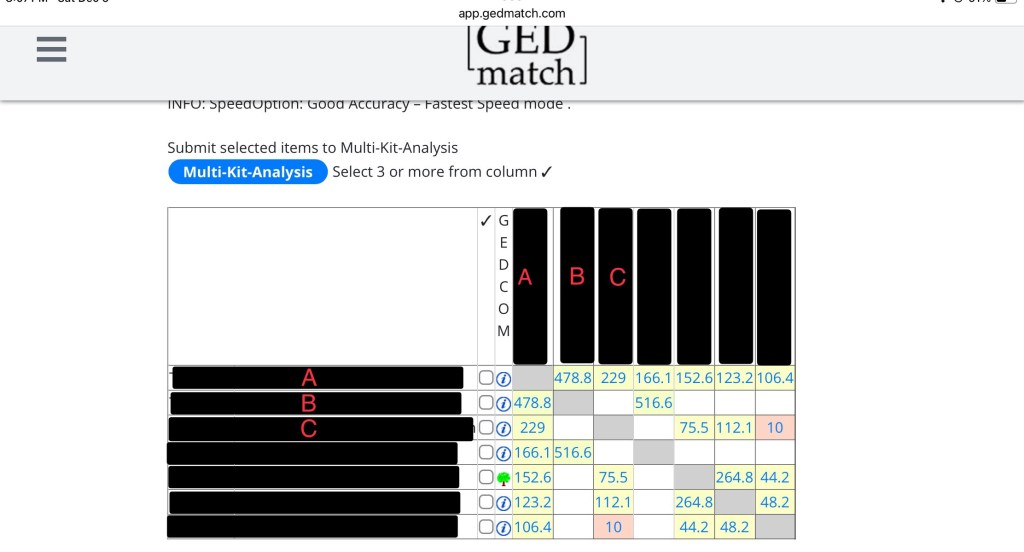
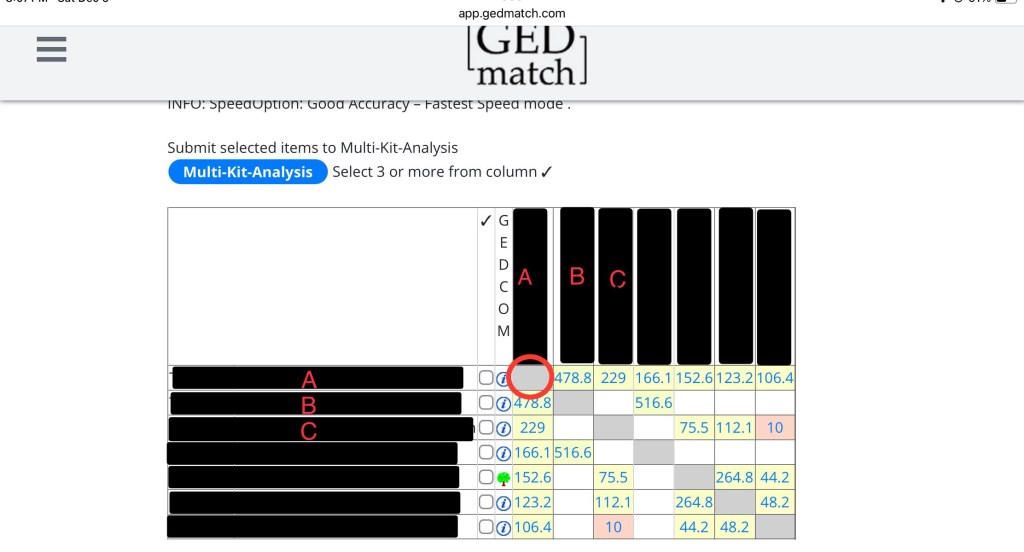
If you don’t know how to read a matrix, here’s an overview. In the picture below, the circle shows you A compared to A. Since it’s the same kit, it’s greyed out.
Next we have two circles, A compared to B and B compared to A. They both have 478.8cM because it’s the same two kits being compared to each other. It’s just two different ways of looking at the same thing. Since this is a fairly close relationship, it’s yellow. The greener the box, the closer the relationship. The further the relationship (like in those boxes with 10), the more red it will be.
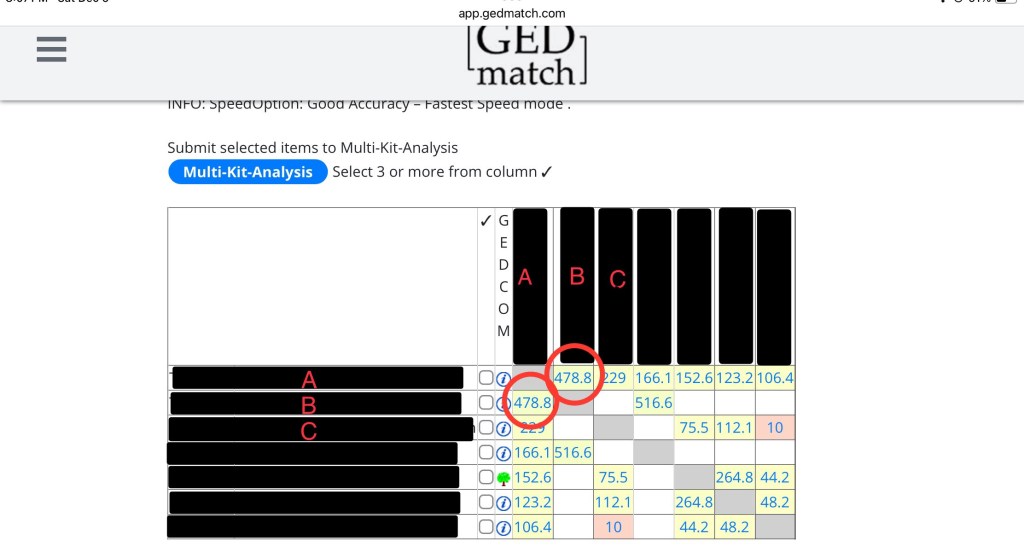
The last thing you need to see is B compared to C. This box is blank! That is because they don’t share any DNA.
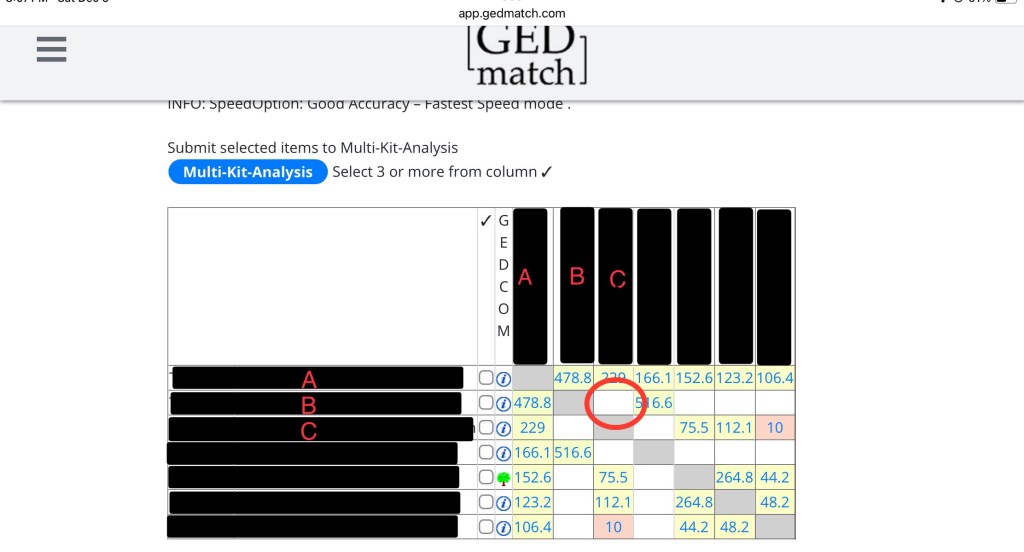
Now you might be wondering, what is the purpose of a matrix? Well, if we know how much DNA everyone shares (or does not share) with everyone else, we can sometimes reconstruct a family tree.
Take this matrix for example:
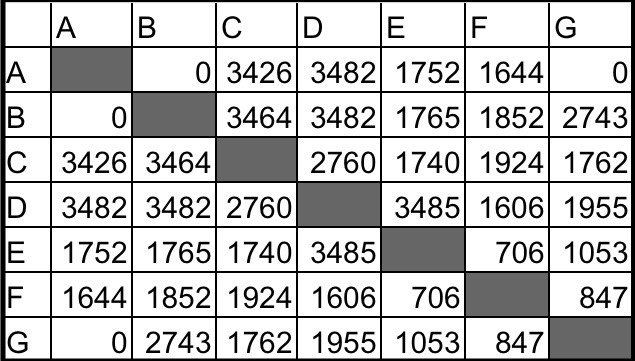
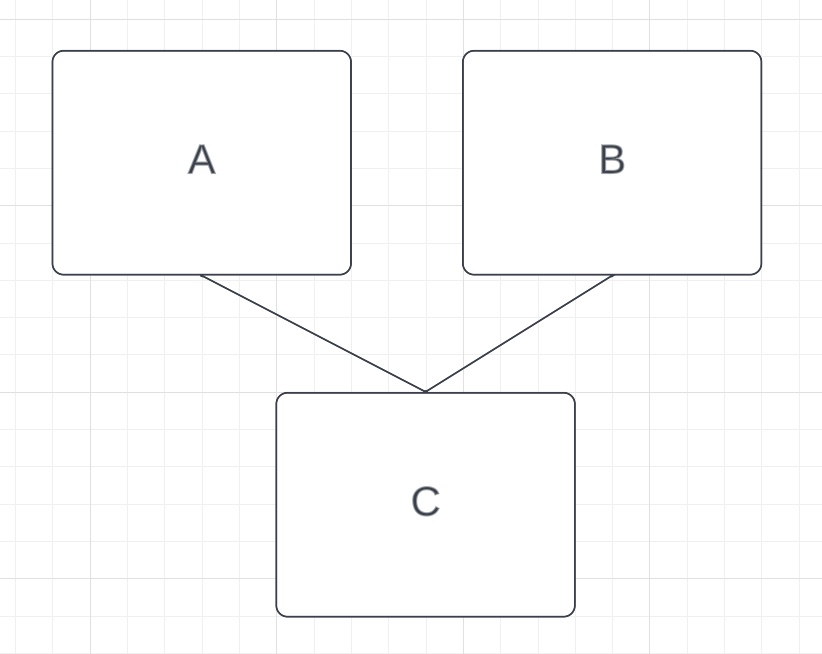
If I were going to build a family tree using this matrix, I would first look at the people that share the most DNA. We know that ~3500cM is a parent-child match, or an identical twin, or a match to oneself. We know that none of these matches are twins or the same person because they share different amounts of DNA with the other matches. For example, if A and C were a twin or the same person they would both share the same amount of DNA with B, but A shares no DNA with B and C shares ~3500 with B. So C has a parent-child match with A and B. Who is the parent and who is the child? If C is the parent of both A and B, then A and B would either be half-siblings or full siblings. Since A and B do not share any DNA, that’s not possible. If A was C’s parent, and B was C’s child, then B would be the grandchild of A, but again, A and B share no DNA, so that’s not possible. The only option that’s left is that C is the child of A and B. We can visualize this (using LucidChart (14), DrawIO (15) or Scapple (16)) like so:
We can also see that A and B both share ~3500cM with D. These are also parent-child matches. The same logic we used for C tells us that D is their child, and this would make C and D full siblings. We can check that the amount of DNA they share, 2760cM, falls within the range for a full sibling at the SharedcM project (18), which it does. Therefore, we have this:
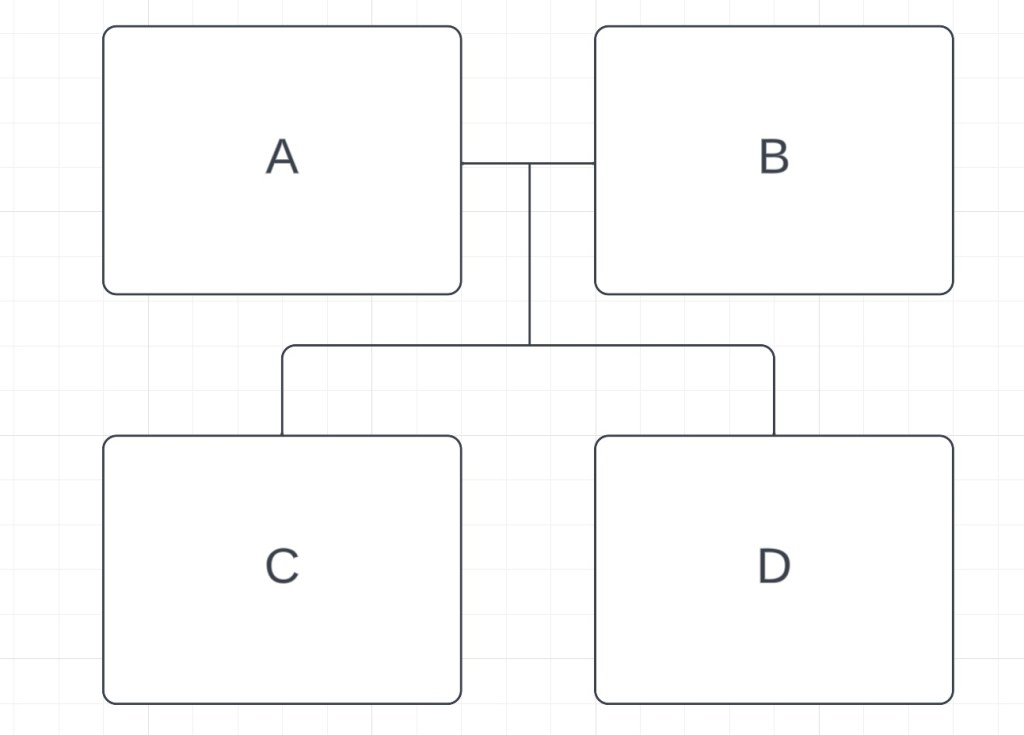
E also shares a parent-child relationship with D. Since D already has two parents, E is D’s child. That means that the amount of DNA C and E share, 1740cM, should fall within the aunt/uncle-niece/nephew range, which it does. E would also be the grandchild of both A and B, and the amount of DNA that E shares with them (~1760cM) also falls into that range. Here’s our family tree so far:
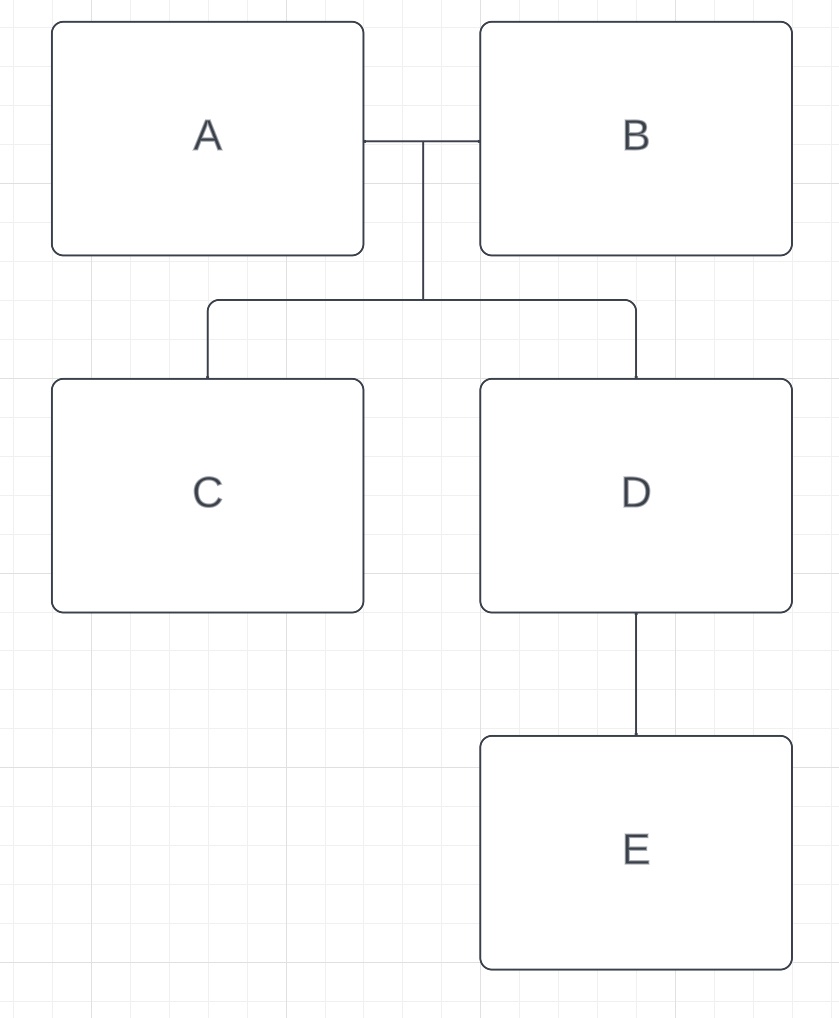
The next highest relationship is between B and G. At 2743cM, G has a full sibling relationship with B, and is not related to A. ~1700-1900cM fits the aunt/uncle relationship with C and D and ~800-1000cM fits the great-uncle/great-aunt relationship with E. We’ll call B and G’s parents Y and Z, and now our tree looks like this:
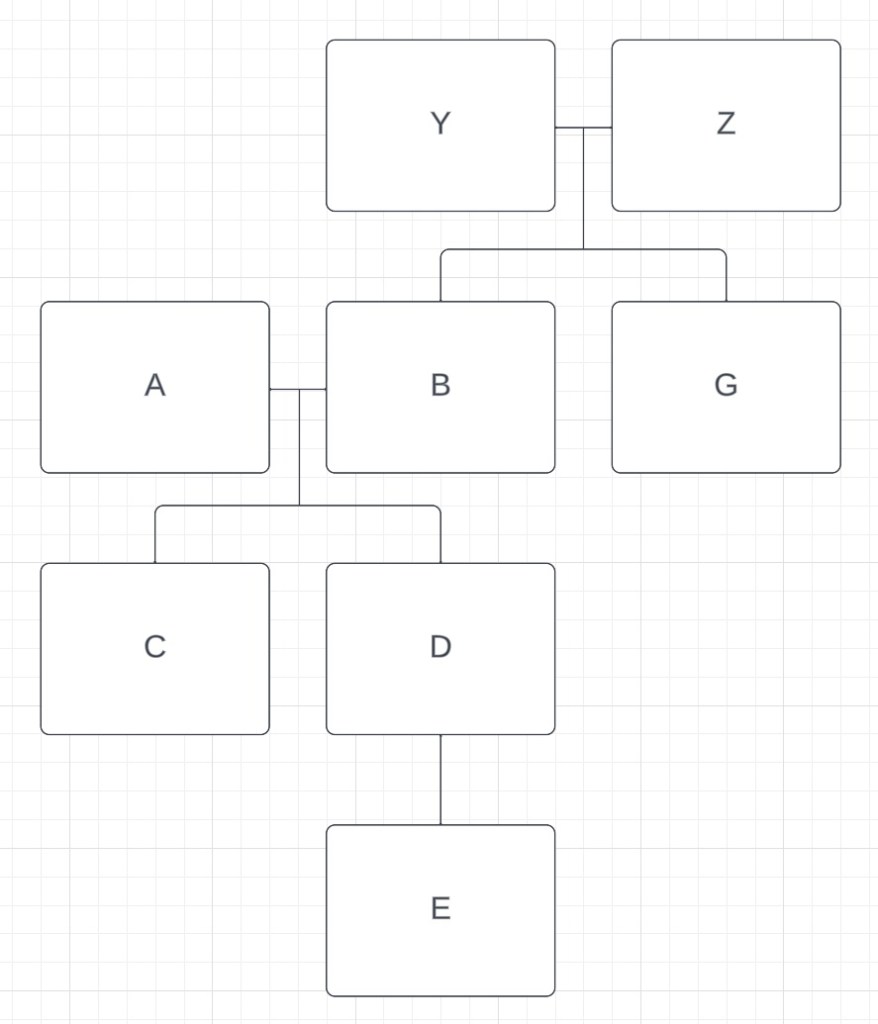
Finally, let’s look at F. The amount of DNA that F shares with A, B, C and D is very similar (~1600-1900cM). The SharedcM project tells us that grandparent-grandchild, aunt/uncle-niece/nephew and half sibling are the most likely relationships. If F was a half sibling to C and D, F would not share DNA with both A and B, only with one of them. It’s possible for F to be a half-sibling to both A and B through different parents, but that would also make F a half-sibling to G, and they share too little DNA for that relationship to be possible. So half-sibling is ruled out. F cannot be the aunt/uncle or niece/nephew to either A or B because they share DNA with both of them, but A and B do not share DNA. But F can definitely be a grandchild of both A and B. There is another full sibling to C and D, one who hasn’t done their DNA. We’ll call them X. If F is the grandchild of A and B, then this would make them the niece/nephew of C and D, and the amount of shared DNA (~1600-1900cM) works for that relationship. Finally, if F is the grandchild of A and B, then E and F are first cousins, and the amount of DNA (706cM) also fits that relationship. Our family tree now includes everyone from our matrix:
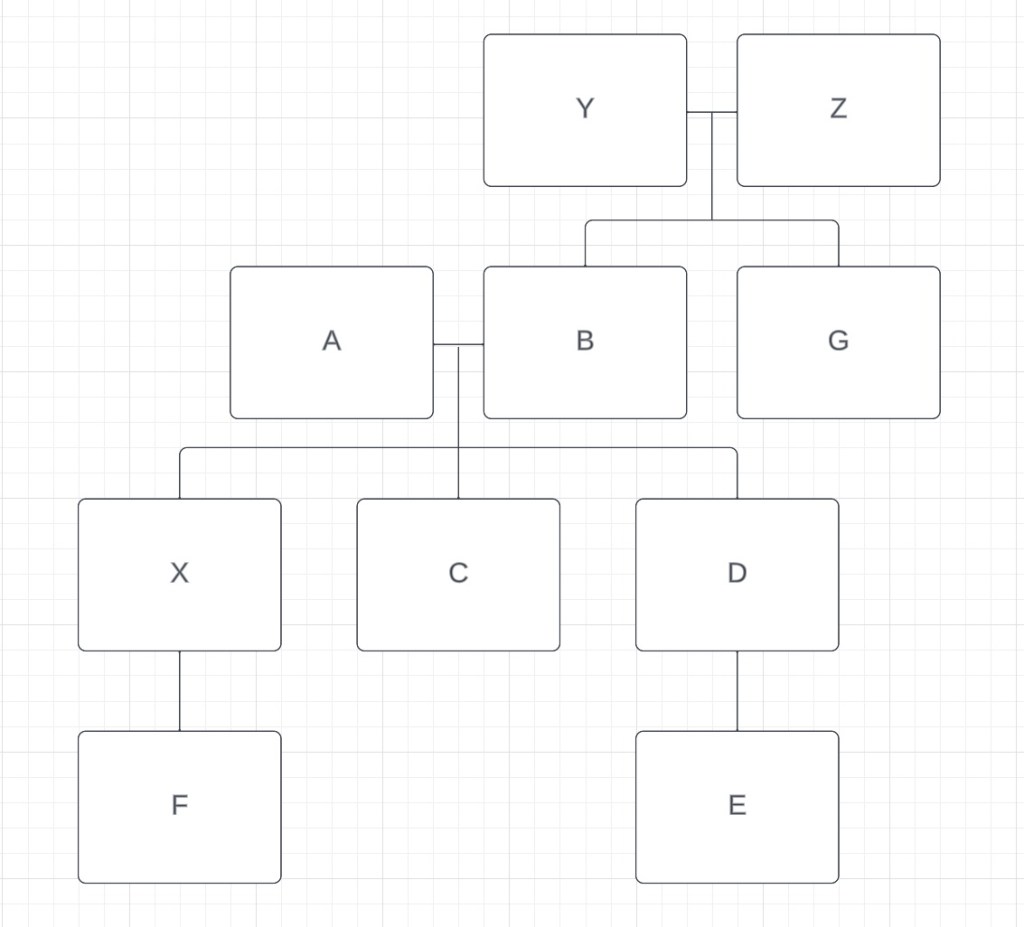
Being able to create a family tree using the amount of shared cM is a useful skill when you don’t know what the relationships are. It’s not always easy, since smaller amounts of shared DNA means more relationship possibilities, but putting families together this way is an essential skill for identifying unknown DNA.
- Gedmatch (https://app.gedmatch.com/login1.php : accessed 12 December 2023).
- Jennifer Wiebe, digital photo, matrix 1, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 2, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 3, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 4 December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 5, December 2023, author’s files.
- Jennifer Wiebe, “Don’t touch the settings!,” Jennealogie, 20 April 2018 (https://jennealogie.com/2018/04/20/dont-touch-the-settings/ : accessed 12 December 2023).
- Jennifer Wiebe, digital photo, matrix 6, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 7, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 8, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 9, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 10, December 2023, author’s files.
- Jennifer Wiebe, digital photo, matrix 11, December 2023, author’s files.
- Jennifer Wiebe, “LucidChart,” Jennealogie, 20 April 2022 (https://jennealogie.com/2022/04/20/lucidchart/ : accessed 12 December 2023).
- Jennifer Wiebe, “Draw.io,” Jennealogie, 29 April 2022 (https://jennealogie.com/2022/04/29/draw-io/ : accessed 12 December 2023).
- Jennifer Wiebe, “Why I Love Scapple,” Jennealogie, 9 July 2020 (https://jennealogie.com/2020/07/09/why-i-love-scapple/ : accessed 12 December 2023).
- Jennifer Wiebe, digital photo, tree 1, December 2023, author’s files.
- “The Shared cM Project 4.0 tool beta v4,” DNA Painter, last updated 26 March 2020 (https://dnapainter.com/tools/sharedcmv4-beta : accessed 12 December 2023).
- Jennifer Wiebe, digital photo, tree 2, December 2023, author’s files.
- Jennifer Wiebe, digital photo, tree 3, December 2023, author’s files.
- Jennifer Wiebe, digital photo, tree 4, December 2023, author’s files.
- Jennifer Wiebe, digital photo, tree 5, December 2023, author’s files.

Leave a comment